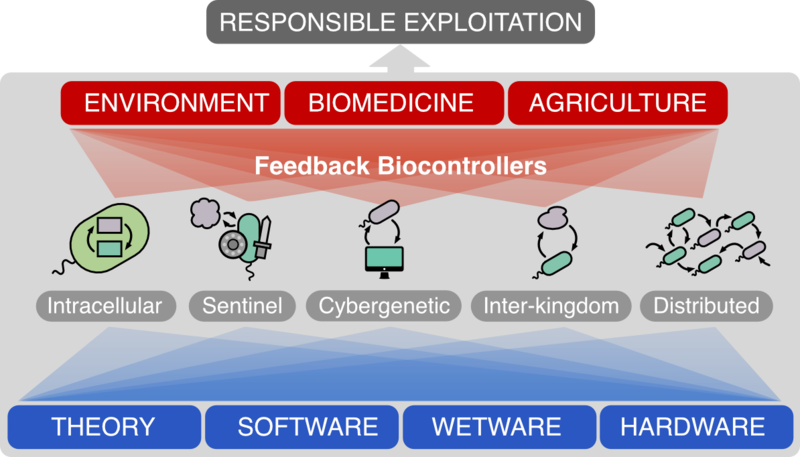
Approach
The robust engineering of biological control systems will be underpinned by the development of four "Engineering Pillars". These cover Theory (mathematical/AI approaches based on systems and control theory to model, design, analyse, and optimise biosystems), Software (computational tools able to translate this theory into conceptual designs), Wetware (experimental methods and biological parts to make designs a biological reality), and Hardware (to comprehensively test, scale-up, and deploy engineered biosystems). Each Pillar feeds directly into an integrated "Design-Build-Test-Learn" cycle rooted in systems and control engineering methods, which will accelerate academic and industrial development of new biotechnologies.
Technologies developed in each Engineering Pillar will be integrated to address outstanding problems in three "Grand Challenge'' application domains: Biomedicine, Agriculture, and the Environment. Our team will work with industrial partners to generate world-leading solutions for each of these areas, demonstrating how biocontrollers can revolutionise scale-up and deployment of reliable engineered biotechnologies.
The long-term goal of our programme grant is to accelerate responsible industrial exploitation, open up the field to other research communities (in the life, medical and social sciences), and support public confidence in the safety and reliability of Engineering Biology.

In this strand we will develop new model-driven design and optimisation tools for predictable and reliable biosystems that operate in uncertain environments. In particular we will design biomolecular feedback control systems for individual and interacting populations of cells and communities as well as machine learning methods and tools for biodesign.

In this strand we will develop new model-driven design and optimisation tools for predictable and reliable biosystems that operate in uncertain environments. In particular we will design biomolecular feedback control systems for individual and interacting populations of cells and communities as well as machine learning methods and tools for biodesign.

The Wetware strand will implement and characterise tuneable biocontrollers that realise various biological feedback architectures. We will develop resource-efficient intracellular biocontrollers that automatically sense and respond to external inputs and can robustify the processes they control against environmental perturbations and disturbances. We will also develop intercellular control architectures that respond to cell-to-cell or interkingdom signalling.
The Hardware strand will develop low-cost high-capability experimental technologies that allow wetware (P3) to be characterised reliably, generating standardised data that feed into the software platform (P2) and ultimately inform theoretical analysis and algorithms (P1). These will include flow reactors for bioprocessing, scalable microenvironments for agricultural applications, and microfluidic control systems.

Integral to addressing climate change and environmental degradation is a shift in the manufacture and processing of chemicals from unsustainable fossil fuels to clean and renewable bio-based alternatives. We will pioneer microbial-based bioproduction systems able to convert low-cost substrates into high-value chemicals in an eco-friendly manner, as well as bioremediation processes to degrade and recycle unwanted by-products.

Reliability and predictability are fundamental requirements for establishing confidence in the safety of new engineered cell-based therapies. C2 will use feedback control implemented over multiple scales (P1) to demonstrate the potential of building new cell-based therapies, with a focus on their integration with microfluidics (P4) to enable safe in vitro testing and scale-up.

Nitrogen fertiliser production and use has a massive energy and environmental cost. Legumes and associated rhizobia bacteria can fix atmospheric nitrogen. However, much of our agricultural productivity depends on cereals that cannot form these useful symbioses. C3 will engineer bacteria-plant symbioses to regulate bacterial association with plants and establish nitrogen fixation and subsequent ammonia release.

This Responsible Exploitation strand will drive forward our vision of a rigorous and systematic approach to Engineering Biology supported by systems and control engineering frameworks, targeting academic and industrial communities, in dialogue with the broader public.

